Researchers: Danielle Posthuma (co-leader), Boudewijn Lelieveldt (co-leader), Martijn van den Heuvel, Sophie van der Sluis, Marcel Reinders, Ahmed Mahfouz, Thomas Höllt.
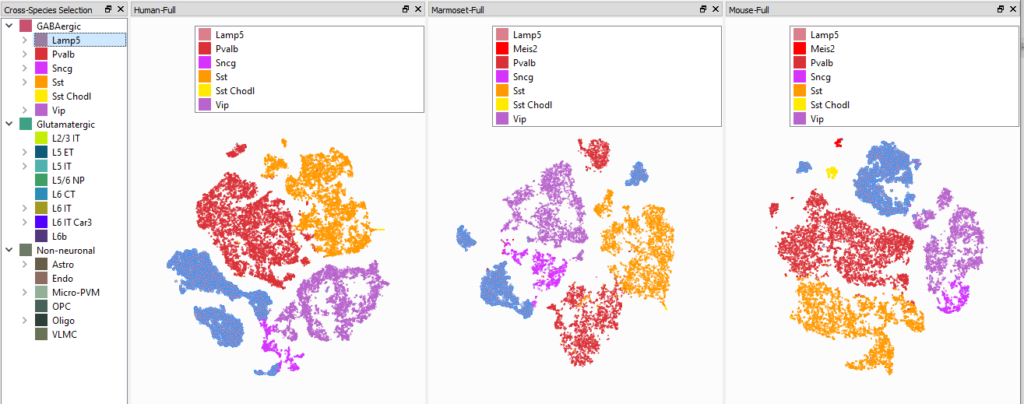
Brainformatic resources are large-scale resources that provide brain-related functional information at the level of genetic variants and genes. The recent availability of such resources has enormous potential for interpreting GWAS results. However, successful biological interpretation of brain-related GWAS results is currently limited by the lack of tools and methods to integrate diverse types of data: molecular (genetic, epigenetic, transcriptomic, and proteomic) and neurobiological scales (cells, circuits, brain regions, and cognitive functions) are now often analyzed separately. A further limitation is also the translation and mapping of data across different species (e.g. how do genes, neural cell types, and network circuits in humans map to those in mouse and vice-versa?).
To overcome these issues WP1 generates novel GWAS discoveries, creates integrated maps of the brain, and develops novel computational tools aimed at annotating GWAS results with brainformatic resources. This approach provides the highly necessary starting points for new functional experiments. WP1 thereby has a fundamental role within Brainscapes for generation of novel biological hypotheses that can be experimentally validated in controlled wet lab environments as part of the research focus of WP2 to WP4, which will generate results that serve as foundation for designing experiments aimed at developing novel treatments in spin-off projects within WP5. Besides data sharing within Brainscapes, novel tools and results from WP1 are made publicly available to the general research community, potentially reaching an even broader genetics and biology field.